We have open postdoctoral positions!
Postdoctoral Applicants are invited to write to Alexei Korennykh:
[email protected]
Mechanism of Metabolic Regulation by the Circadian Protein Nocturnin
Our work is now published on Nature Communications:
https://www.nature.com/articles/s41467-019-10125-z
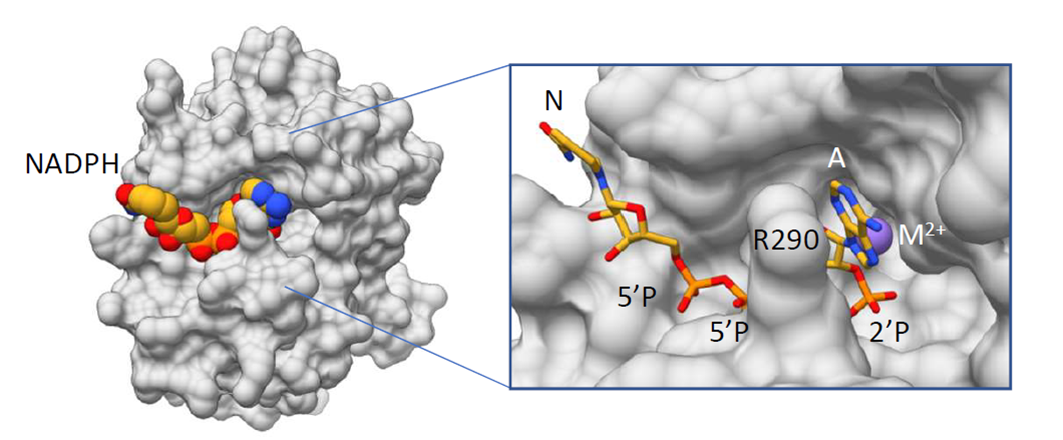
NADP(H) is the substrate of Nocturnin
About 20 years ago a wonderful discovery was made by Carla Green and Joseph Besharse, who showed that the protein Nocturnin is controlled by the circadian clock.
Nocturnin was later found to be important for metabolism and control of obesity and these pioneer works sparked off a considerable interest in Nocturnin. Previous studies indicated that Nocturnin might deadenylate mRNA in vitro. Regulation of metabolism is therefore attributed to a globally altered stability of metabolic enzyme mRNAs. However, our laboratory recently encountered a puzzling lack of deadenylase activity in highly purified Nocturnin (https://www.nature.com/articles/s41598-018-34615-0). The same conclusion was independently made by another laboratory, also in 2018 (https://academic.oup.com/nar/article/46/12/6257/5026266).
We took an unbiased approach to identify the possible non-RNA substrate of Nocturnin, and found that Nocturnin catalyzes the removal of 2’-phosphate from NADP+ and NADPH, converting them to NAD+ and NADH. The involvement of Nocturnin in metabolism has been widely recognized, but it had not been expected that the enzyme directly targets the central cofactors, NADPH and NADP+, in anabolic and catabolic reactions.
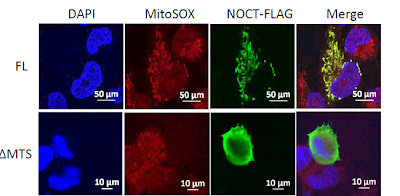
Further, we show for the first time that Nocturnin localizes to the mitochondrion.
Discovery of the mechanism for Drosophila Curled, a hallmark gene in fly genetics with 100 years of history
The first description of Nocturnin took place more than a hundred years ago in the work of Thomas Hunt Morgan. While working with Drosophila, Thomas Hunt Morgan encountered a mutation he called Curled (cu) because it caused a peculiar upward wing curvature. The cu mutant has become a broadly used marker in fruit fly genetics and the curled phenotype has been linked to a defect in metabolism. Sequence analysis by S. Gronke et al. (Genetics 2009) showed that Curled is a homologue of Nocturnin.
Since this result, Curled was also classified as an mRNA deadenylase and its biologic effects were attributed to mRNA decay.
Our data suggest that Curled from Drosophila has the same activity as human Nocturnin and cleaves NADPH.
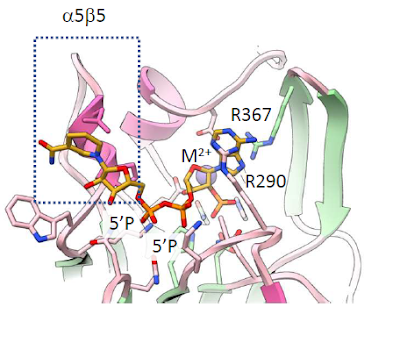
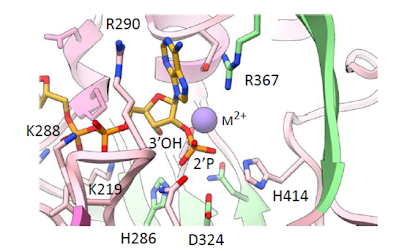
2.7Å X-ray structure
We solved the crystal structure of human Nocturnin with NADPH bound. Our structure explains why Nocturnin active site does not cleave RNA, and shows also how it selectively cleaves only NADP(H).
A new enzyme class
Nocturnin has Enzyme Commission classification 3.1.13.4: poly(A)-specific 3'-exoribonuclease. We show that instead Nocturnin is a NADP(H) 2’-phosphatase.
In the moment there is only one example of a NADP(H) 2’-phosphatase, MESH1, described in a preprint publication on BioRxiv (https://www.biorxiv.org/content/early/2018/05/17/325266).
MESH1 has a different structure than Nocturnin, it does not localize to the mitochondria, and in contrast to Nocturnin/Curled, it does not have well established biologic roles. However, the discovery of two structurally and biologically distinct 2’-phosphatases calls for the creation of an Enzyme Commission entry for this novel enzyme class.Immune & Stress Response Mediated by RNA
We seek a precise molecular understanding of how coding and non-coding RNAs facilitate mammalian stress responses. In particular, we focus on stress-activated RNA cleavage mechanisms, which regulate gene expression and control cell fate to eliminate damaged/overwhelmed cells.
Human cells activate these mechanisms in response to a strong mammalian immunogen - double-stranded RNA (dsRNA). We study the molecular machinery involved in this cellular program by X-ray crystallography, biochemistry and biophysics. This work goes hand in hand with cell biology and genomics projects in the laboratory, which employ RNA-seq and custom RNA-seq methods.
Our ultimate goal is to gain fundamental knowledge with clear biomedical implications, which would elucidate the roles of coding and non-coding RNAs in pathogen defense, interferon and inflammatory signaling and autoimmune diseases.
Keywords: Interferon, inflammation, dsRNA, tRNA, Y-RNA, t-RNA fragments, tRFs, Y-RNA fragments, YRFs, autoantigen, RNase L, oligoadenylate, 2-5A, OAS1, OAS3, microRNA, adhesion, apoptosis, lupus, metastasis, EMT.
Structural biology
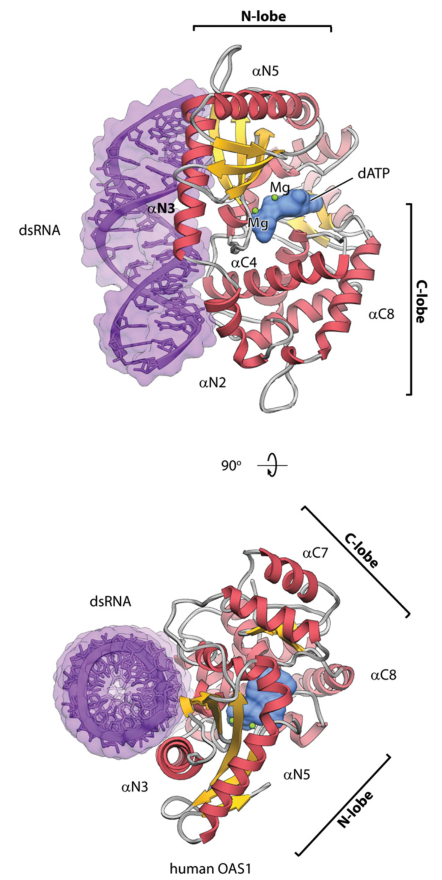
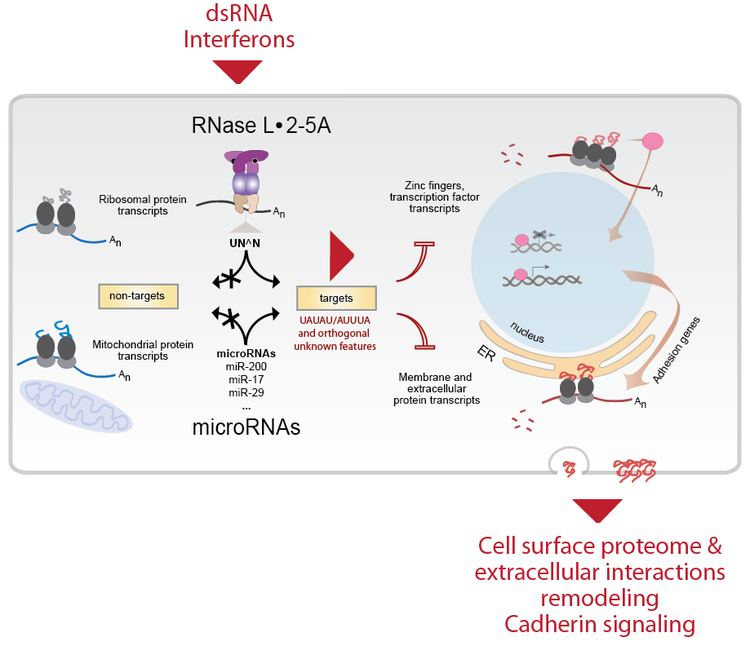
Receptors of dsRNA. Our work is providing structural and biochemical insights into double-stranded RNA (dsRNA) sensors, which activate mRNA decay by producing a signaling RNA-like molecule "2-5A" with unusual 2',5'-linkages. The dsRNAs molecules are not abundant in normal cells, but they become abundant upon viral infections or DNA damage that presumably strips off repressive DNA methylation marks from endogenous retrovirus sites. The endogenous dsRNA production, surveillance and physiologic roles are very intriguing and not fully understood.
STRUCTURE OF HUMAN OAS1 WITH dsRNA
HUMAN OAS3 IS A SENSOR OF LONG dsRNA
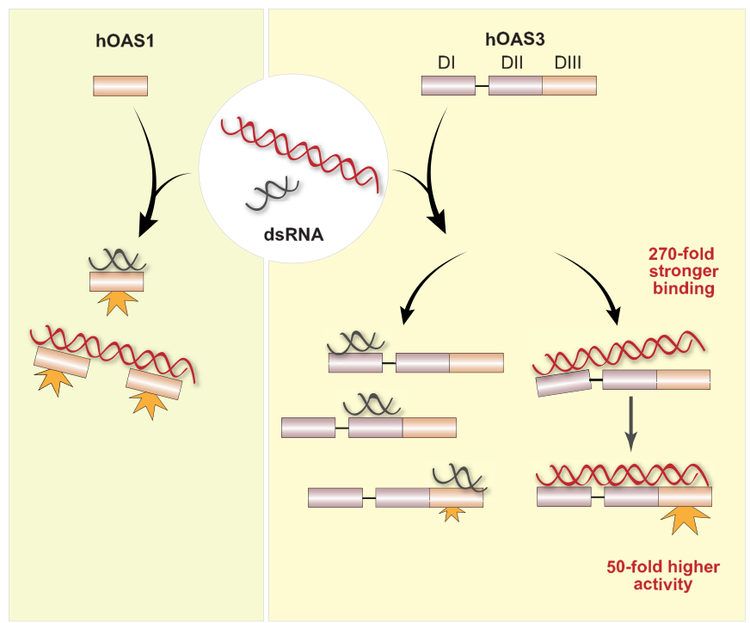
Human cells express three related but different sensors of dsRNA that synthesize 2-5A. Using X-ray crystallography and biochemistry we determined that the largest OAS family member, OAS3, is a selective sensor of long dsRNA. OAS3 contains three duplicated domains. The C-terminal domain (DIII) is catalytically active and makes 2-5A, whereas the N-terminal domain (DI) is catalytically inactive and serves for high-affinity dsRNA recruitment and for measurement of minimal dsRNA length.
Donovan J, Whitney G, Rath S, Korennykh A.
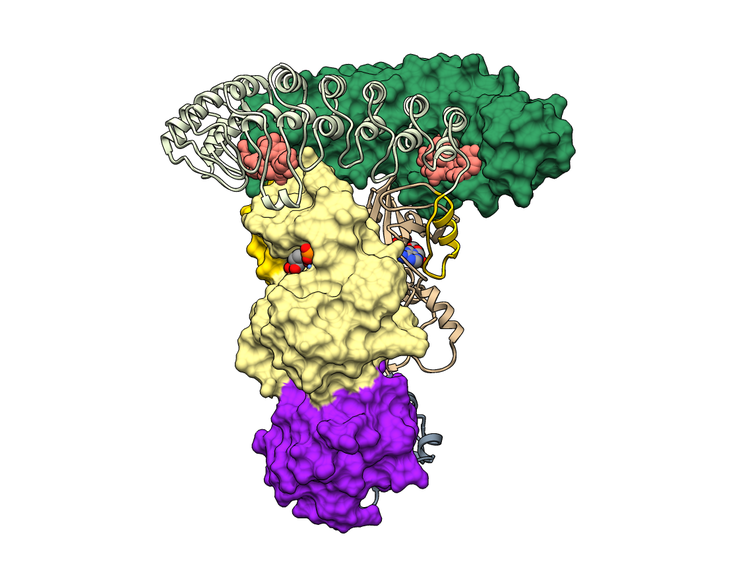
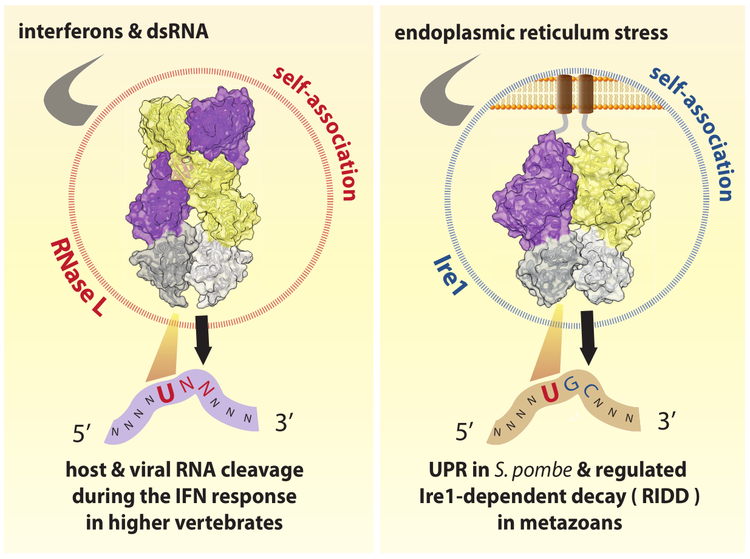
DsRNA-induced mRNA decay. We are also learning about the 2-5A receptor, RNase L. RNase L contains a protein kinase domain and belongs to the family of more than 500 mammalian protein kinases. RNase L does not phosphorylate any known proteins, which makes it different from most kinases. Uniquely, RNase L cleaves cellular RNAs during the interferon response, which protects the cells from viruses and bacteria, and blocks cell proliferation. We solved the crystal structure of human RNase L with bound 2-5A and RNA substrate, which explains how this enzyme is regulated.
STRUCTURE OF HUMAN RNASE L WITH 2-5A AND RNA SUBSTRATE
Han Y, Donovan J, Rath S, Whitney G, Chitrakar A, Korennykh A.
Han Y, Whitney G, Donovan J, Korennykh A.
Cell biology & RNA-seq
Another area of our work is understanding the precise mechanism and roles of mRNA cleavage by RNase L and by other immune proteins. It has been long known that RNase L is activated during the IFN response and that it can inhibit viral and bacterial infections, and cell proliferation. However, mapping the cellular mRNA targets that RNase L cleaves to achieve these effects proved challenging. We used a combination of RNA-seq strategies to identify the mRNA targets of RNase L and map its cellular program.
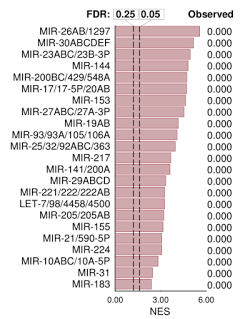
We found that RNase L cleaves the same mRNAs as are targeted by the microRNA miR-200. MiR-200 is a remarkable microRNA because it is a master regulator of cell adhesion, proliferation and is one of the key modulators of epithelial-to-mesenchymal transition (EMT) during metastasis. Thus, RNase L is functionally a mimic of miR-200. RNase L is unique in that unlike miR-200, it is activated downstream of IFN and dsRNA signaling. RNase L could be approximately described as an "on-demand miR-200 in the innate immune system".
As expected from our analogy, RNase L and miR-200 have many similar effects on human cells. Both block proliferation, both block adhesion and both target ZEB1 transcription factor and regulate cadherins.
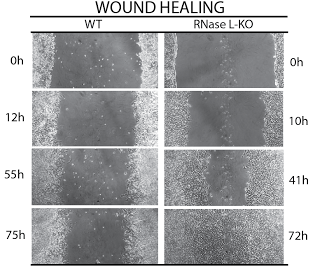
RNA-SEQ OF RNASE L TARGETS IN T47D HUMAN BREAST CANCER CELLS

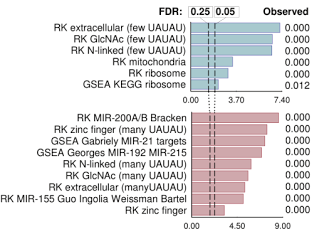
TargetScan 6 gene signatures
THE EMERGING PATHWAY OF RNASE L
KO = knockouts
Rath S, Donovan J, Whitney G, Chitrakar A, Wang W, Korennykh A.
Some of our main techniques
Our work is funded by Princeton University, Sidney Kimmel Foundation, Burroughs Wellcome Foundation, and NIH.
Recent Publications
Contact
Korennykh Lab
Department of Molecular Biology
216 Schultz Laboratory
Washington Road
Princeton, NJ 08544
p 609-258-7303
f 609-258-6730
Faculty Assistant
Karen Plantarich
217 Schultz Laboratory
[email protected]
p 609-258-5028
Lab Website
molbiolabs.princeton.edu/korennykh